Research
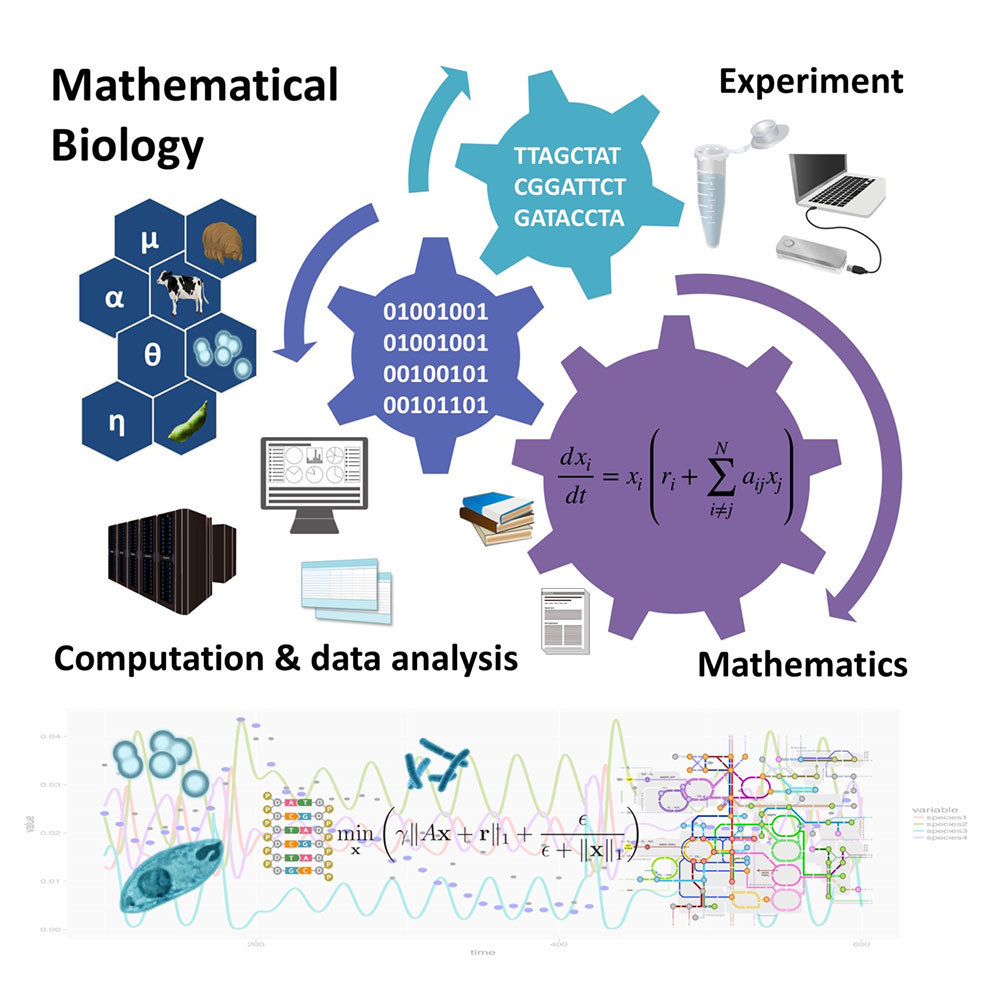
In our laboratory, mathematical and statistical models are employed to work with experimental or field biologists in order to understand biological phenomena of interest, or quantitatively characterize data. Primary research target of our laboratory is to develop ecomics and ecomimetics. The former represents a type of research to investigate a given microbial community by integrating omics data on the basis of community ecology theories and methodologies. While the latter is motivated by biomietics to represent a type of study to extract the essence of community assembly rules from existing microbial ecosystem to construct a novel artificial microbial community. Other related topics in our laboratory include omics data analysis in precision medicine or development of novel mathematical tools for data analysis in life sciences.
Mathematical modeling of microbial community dynamics: understanding community assembly rules in microbial societies
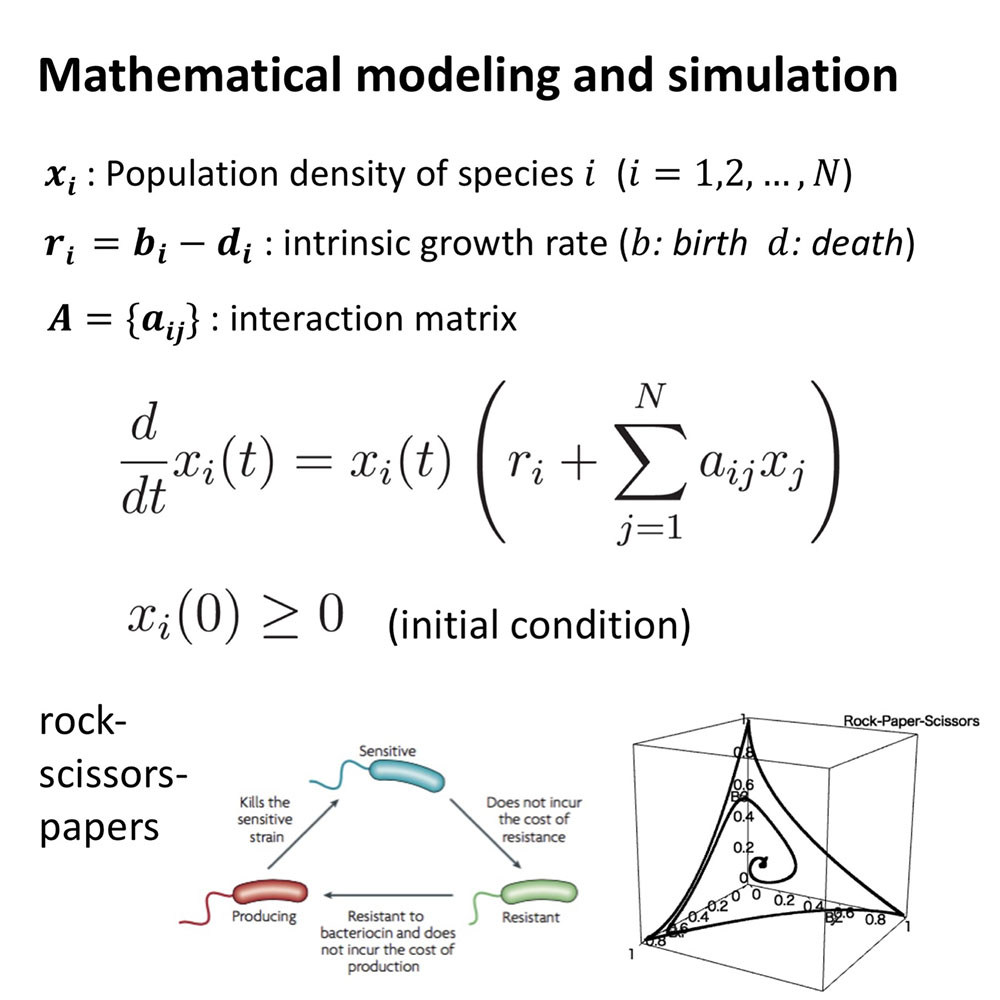
A microbial community changes dynamically as a result of complex interactions among ingredients such as bacteria and virus. Mathematical modeling is useful to constitutively understand dynamical process of a microbial community in terms of microscopic community assembly rules. The purpose of this subtopic is to understand the role of core interactions on dysbiosis, compositional change of a community during disease progression toward reducing species diversity.
Quantitative data analysis of microbial community dynamics: mining community assembly rules in microbial societies
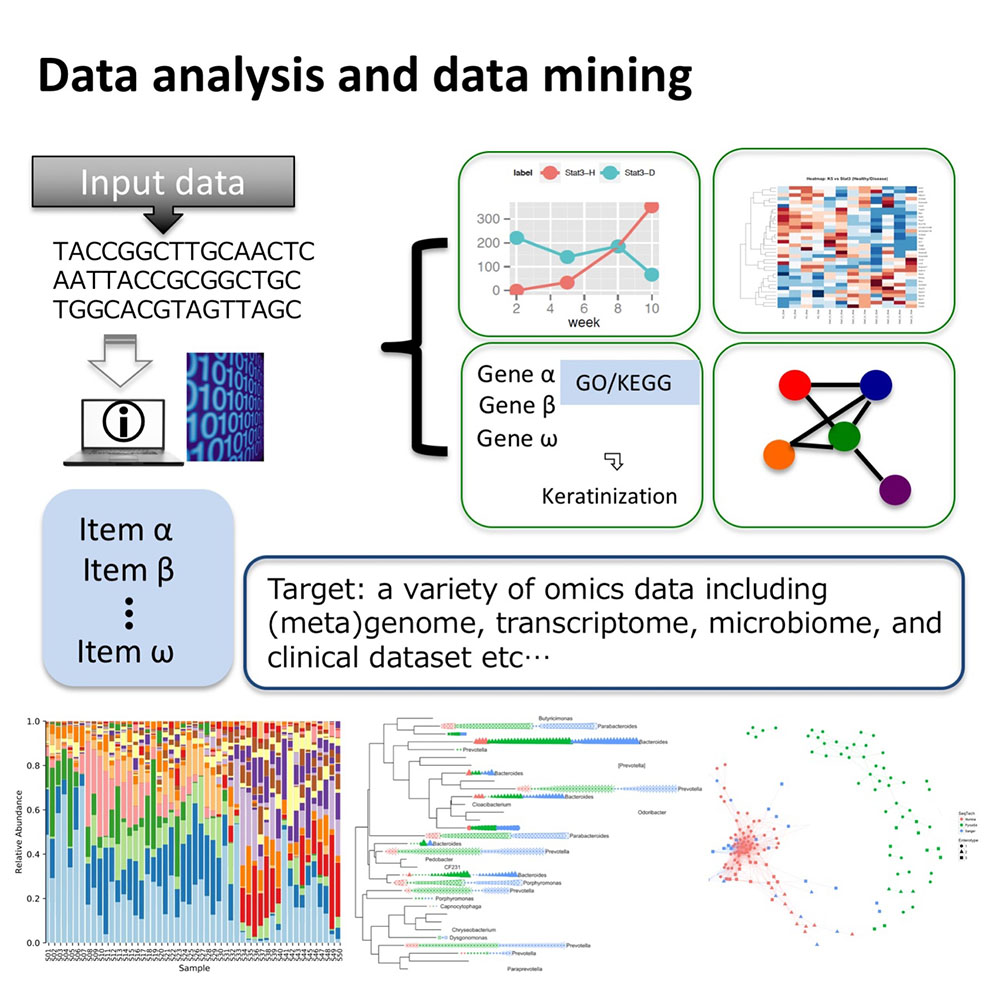
Data analysis is indispensable to identify ingredients that drive dynamic change of a microbial community. The purpose of this subtopic is to apply various methodologies in data science for extracting critical microscopic factors and community assembly rules that constitute a microbial community. Processing environmental metagenomics data is a primary step to highlight core interactions mediating disease progression associated with dysbiosis.




