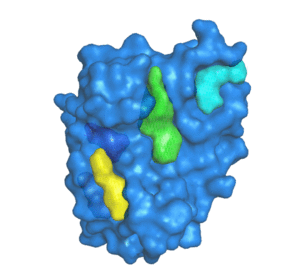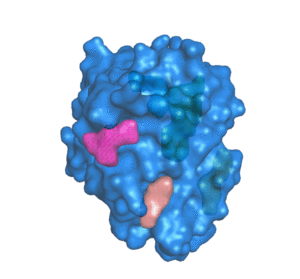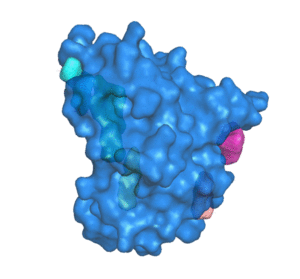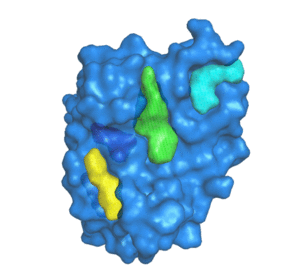
PDBID: 1ION
5 pockets detected.
POCASA (POcket-CAvity Search Application) is an automatic program that implements the algorithm named Roll which can predict binding sites by detecting pockets and cavities of proteins of known 3D structure.
First, a 3D grid system is created and filled with atoms in the protein molecule. Second, a probe sphere is adopted to roll along the protein surface to generate a "probe surface" based on the inner border tracing algorithm in the image processing field. Then, the regions between the protein and probe surface or those surrounded by the protein surface are defined as pockets and cavities, respectively. To remove noise points, two parameters were designed: Single-Point Flag (SPF) and Protein-Depth Flag (PDF). Moreover, POCASA can predict pockets differing in shape and volume by adjusting the radius of the probe sphere. Finally, Volume-Depth (VD) quantitively describing the volume and position information of pockets was designed as a pocket-ranking descriptor.

PDBID: 1ION
5 pockets detected.