
ABSTRACT
SusB, an 84-kDa α-glucoside hydrolase involved in the starch utilization system (sus) of Bacteroides thetaiotaomicron, belongs to glycoside hydrolase (GH) family 97. We have determined the enzymatic characteristics and the crystal structures in free and acarbose-bound form at 1.6A resolution. SusB hydrolyzes the α-glucosidic linkage, with inversion of anomeric configuration liberating the β-anomer of glucose as the reaction product. The substrate specificity of SusB, hydrolyzing not only α-1,4-glucosidic linkages but also α-1,6-,α-1,3-, and α-1,2-glucosidic linkages, is clearly different from other well known glucoamylases belonging to GH15. The structure of SusB was solved by the single-wavelength anomalous diffraction method with sulfur atoms as anomalous scatterers using an in-house x-ray source. SusB includes three domains as follows: the N-terminal, catalytic, and C-terminal domains. The structure of the SusB-acarbose complex shows a constellation of carboxyl groups at the catalytic center; Glu532 is positioned to provide protonic assistance to leaving group departure, with Glu439 and Glu508 both positioned to provide base-catalyzed assistance for inverting nucleophilic attack by water. A structural comparison with other glycoside hydrolases revealed significant similarity between the catalytic domain of SusB and those of α-retaining glycoside hydrolases belonging to GH27, -36, and -31 despite the differences in catalytic mechanism. SusB and the other retaining enzymes appear to have diverged from a common ancestor and individually acquired the functional carboxyl groups during the process of evolution. Furthermore, sequence comparison of the active site based on the structure of SusB indicated that GH97 included both retaining and inverting enzymes.
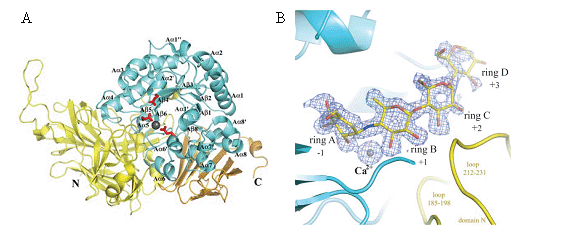
FIGURE CAPTION
(A) Overall structure of SusB in apo and acrabose complex form.
Three catalytic residues, Glu439, Glu508, and Glu532, are shown as stick models in red,
and a bound calcium ion is shown as a gray sphere. Monomer structure of SusB.
Domains N, A, and C are shown in yellow, cyan, and gold, respectively.
The secondary structure elements of domain A are labeled following the order of typical (β/α)8 barrel structures,
and the prime and double prime refer to the atypical elements.
(B) Composite of the acarbose molecule and a calcium ion, and their omit map.
The contour level of the Fo - Fc map is 3σ. Domains N and A are shown in yellow and cyan, respectively.
REFERENCES
Structural and Functional Analysis of a Glycoside Hydrolase Family 97 Enzyme from Bacteroides thetaiotaomicron.
Momoyo Kitamura, Masayuki Okuyama, Fumiko Tanzawa, Haruhide Mori, Yu Kitago, Nobuhisa Watanabe, Atsuo Kimura, Isao Tanaka and Min Yao.
J. Biol. Chem. 283, 36328-36337 (2008)