The RtcB protein has recently been identified as a 3'-phosphate RNA ligase that directly joins an RNA strand ending with a 2',3'-cyclic phosphate to the 5'-hydroxyl group of another RNA strand in a GTP/Mn2+-dependent reaction. Here, we report two crystal structures of Pyrococcus horikoshii RNA-splicing ligase RtcB in complex with Mn2+ alone (RtcB/Mn2+) and together with a covalently bound GMP (RtcB-GMP/Mn2+). The RtcB/Mn2+ structure (at 1.6 Å resolution) shows two Mn2+ ions at the active site, and an array of sulfate ions nearby that indicate the binding sites of the RNA phosphate backbone. The structure of the RtcB-GMP/Mn2+ complex (at 2.3 Å resolution) reveals the detailed geometry of guanylylation of histidine 404. The critical roles of the key residues involved in the binding of the two Mn2+ ions, the four sulfates, and GMP are validated in extensive mutagenesis and biochemical experiments, which also provide a thorough characterization for the three steps of the RtcB ligation pathway: (i) guanylylation of the enzyme, (ii) guanylyl-transfer to the RNA substrate, and (iii) overall ligation. These results demonstrate that the enzyme's substrate-induced GTP binding site and the putative reactive RNA ends are in the vicinity of the binuclear Mn2+ active center, which provides detailed insight into how the enzyme-bound GMP is transferred to the 3'-phosphate of the RNA substrate for activation and subsequent nucleophilic attack by the 5'-hydroxyl of the second RNA substrate, resulting in the ligated product and release of GMP.

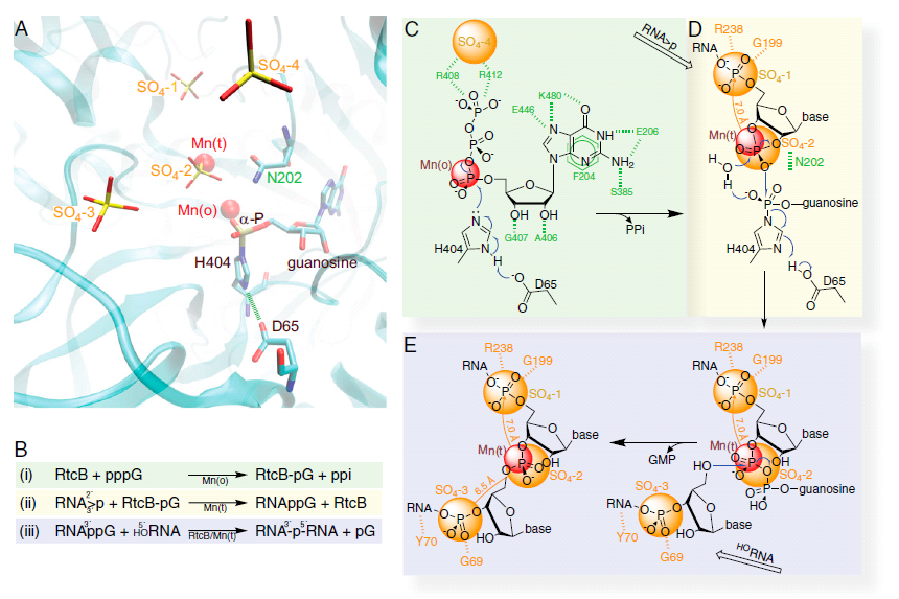
An overview of structure-derived reaction mechanism of the 3'-P RNA ligation. (A) Composite features of the two PHRtcB structures reported here with emphasis on the relationship of four sulfates and two Mn2+ ions with guanylylated H404 in the catalytic site in ribbons representation along with other highlighted residues, namely D65 and N202. (B) A summary of the RtcB-catalyzed pathway in three individual steps (described in the main text and schematized in C-E) as deduced from our structures and biochemistry. (C) The RtcB residues that specifically recognize the substrate GTP are shown in green. The putative PPi site coincides with SO4-4 (orange). The carboxylate group of D65 is hydrogen bonded to H404 Nε1-H, thus enhancing the electron negativity of H404 Nδ2 for nucleophilic attack on GTP's α-phosphate that is in contact with the Mn2+ in octahedral geometry. (D) Coordination of tetrahedral Mn2+ allows binding the RNA strand ending with the 2', 3'-cyclic phosphate (RNA>p) superimposed with the last two phosphates at the SO4-1 and SO4-2 sites. (E) Positioning the HORNA strand with its first phosphate at the SO4-3 allows the optimal geometry of the 5'-hydroxyl for nucleophilic attack on the 3'-P activated terminus to form the internucleotide phosphodiester bond with the release of GMP.